In vitro characterization and rational analog design of a novel inhibitor of telomerase assembly in MDA MB 231 breast cancer cell line
Novel ligand generation, docking, and chemical diversity analysis
Check the publication
In this work, I generated a library of 100 novel derivatives from a lead compound. Afterwards, the generated compounds were converted to 3D formats using industry standard Python packages, and were analyzed using the Protein-Ligand Interaction Profiler software. Afterwards, a Docking-Based Hight Throughput Virtual Screening was performed using the novel ligands.
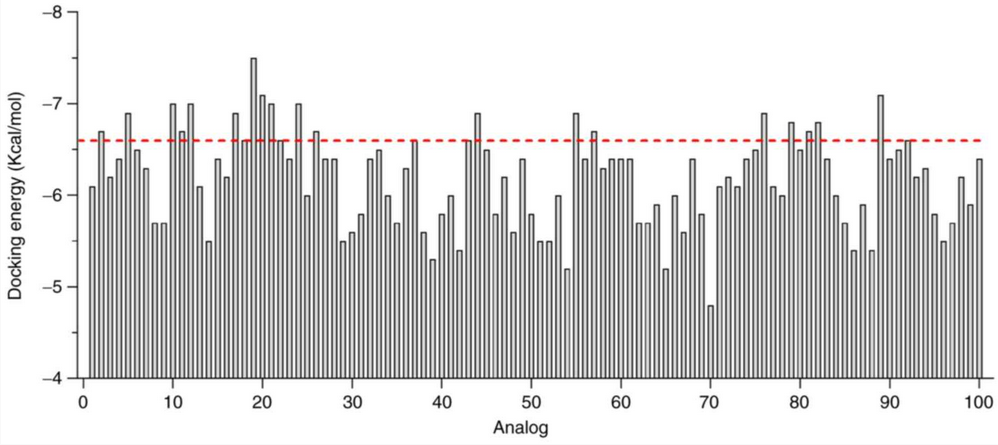
The figure shows the binding affinity of each novel derivative, with the red line showing the original compound affinity.
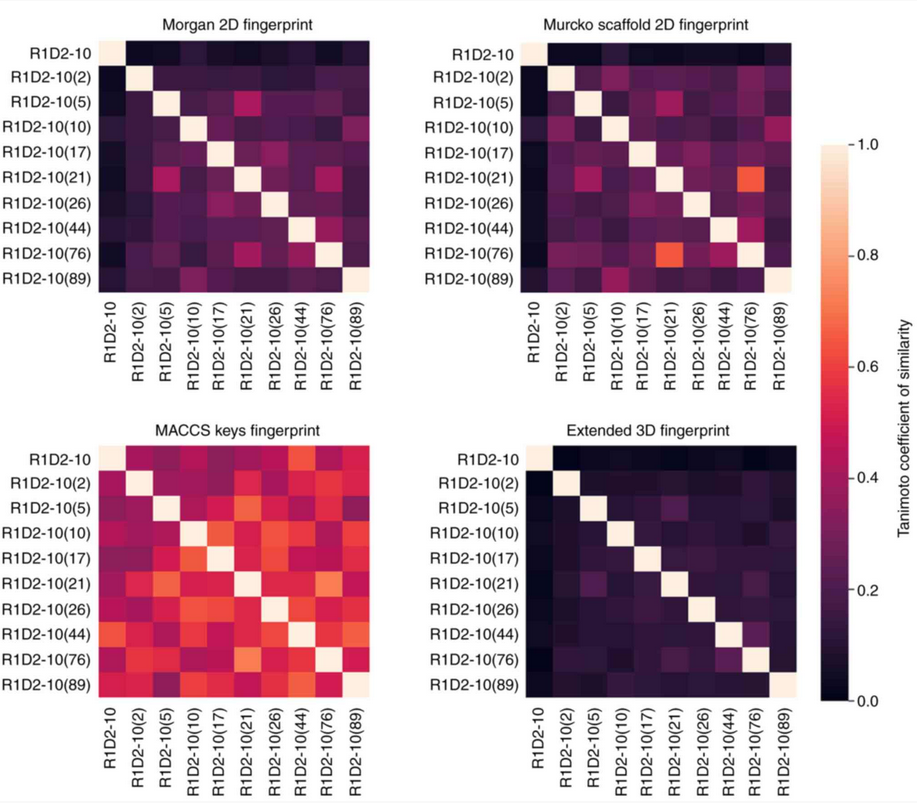
After filtering out unwated compounds with adverse effects, I encoded the different chemical features of the selected compounds in bit vectors that I later compared using the Tanimoto Coefficient of Similarity. This resulted in four different similarity matrices, each evaluating a different property of the molecules, that can be seen below.