Rational design of PIN1 inhibitors for cancer treatment based on conformational diversity analysis and docking based virtual screening
Conformational diversity and molecular dynamics analysis of PIN1
Check the publication
In this work, I analyzed 42 conformations of the PIN1 protein. To search for molecules that would bind to the whole range of conformations, I compared searched for the maximum RMSD pair, and evaluated the Z-score of the per-residue RMSD. This way, we confirmed that the selected region for the screening corresponds to a relatively stable region of the protein, thus making it a good target for a High Throughput Virtual Screening campaign.
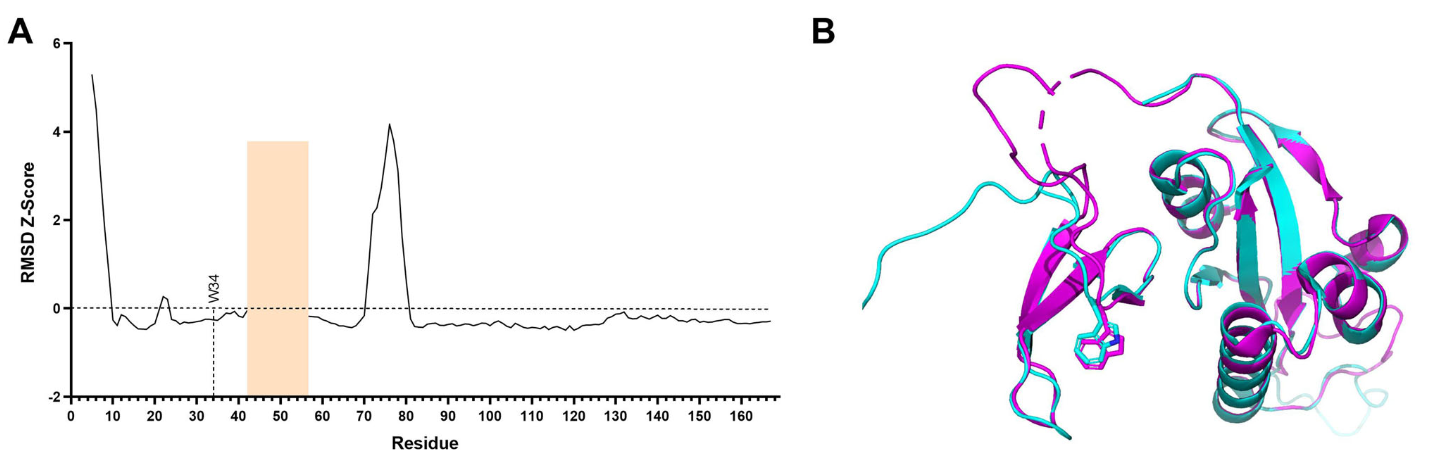
Figure A shows that the residue W34 and it’s surroundings are below 0 for RMSD Z-score, meaning they are relatively stable, thus they were targeted for the HTVS. Figure B shows a supperposition of the protein structures 1F8A-B and 2F21-A, the maximum RMSD pair for PIN1 conformations.
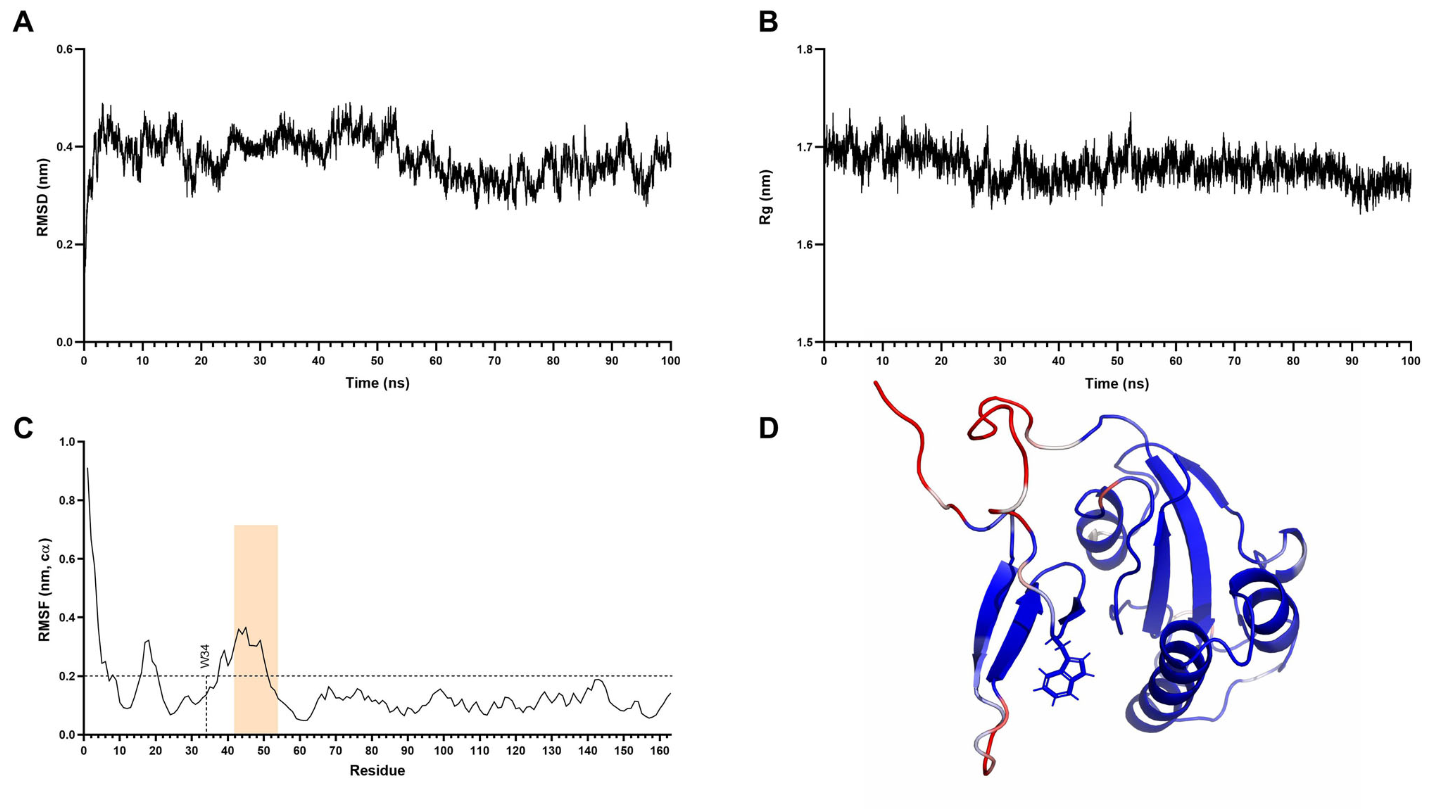
To corroborate the previous result, I performed a 100ns molecular dynamic simulation, which simulates the movements of the protein. As a result, I was able to calculate the Root Mean Square Fluctuation per residue of the protein. The RMSF, similarly to the RMSD, measures the mobility of the residues, but along the course of the simulation. As can be seen in the figure below, the residue W34 and its sourroundings are stable along the simulation, confirming the previous result! Figure A and B show the stability of the system, while figure C shows the RMSF. Figure D is a 3D structure of PIN1 colored by the RMSF value of the residue. W34 can be seen in blue, being the only residue in the sticks representation. The RMSF color code in this case is ‘blue < white < red’